EternaBot: RNA Secondary Structure Design
EternaBot is an efficient algorithm for determining a RNA sequence for a given secondary structure, or base pairing arrangment. It was the culmination of years of work from hundreds of players of the Eterna cloudlab project. Try it for yourself below!
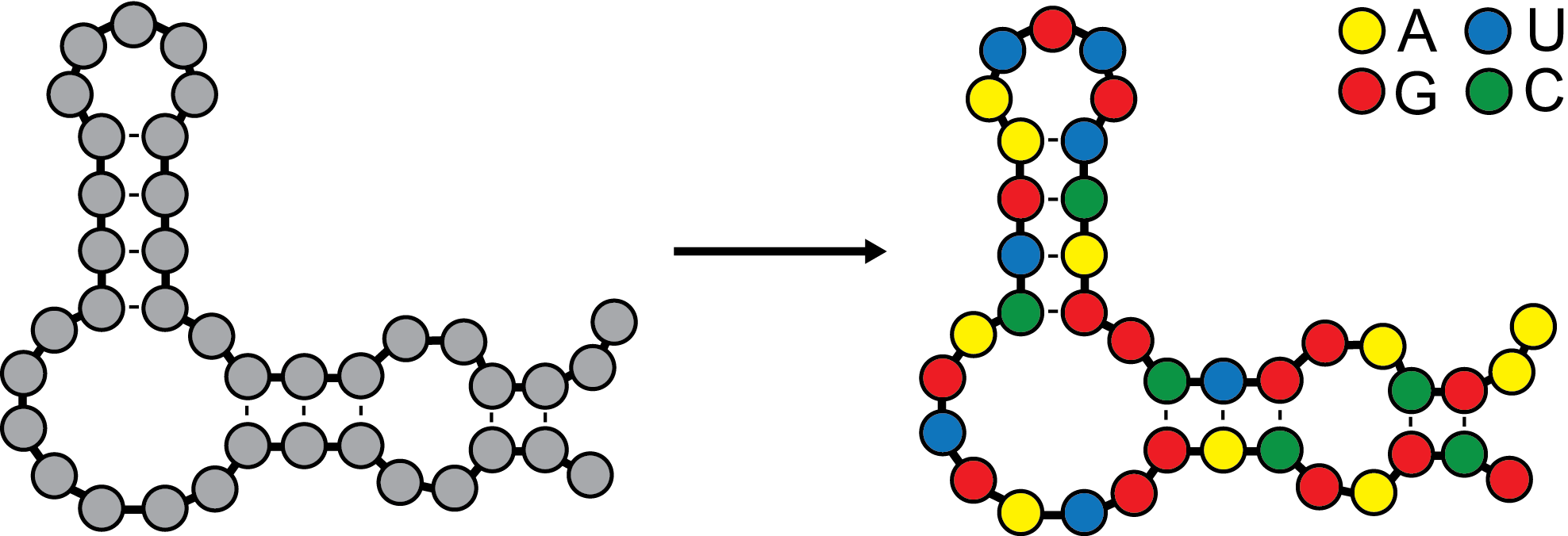

Submit your EternaBot job
To limit how long it takes to run jobs, this webserver has a limit of 200 nucleotides and 10 solutions. For larger jobs, please use a local copy of Eternabot. Don't know where to start? Try an example input secondary structure and sequence.